Studienarbeit: MVCNN
Multiview classification of chloroplast cells
Abstract:
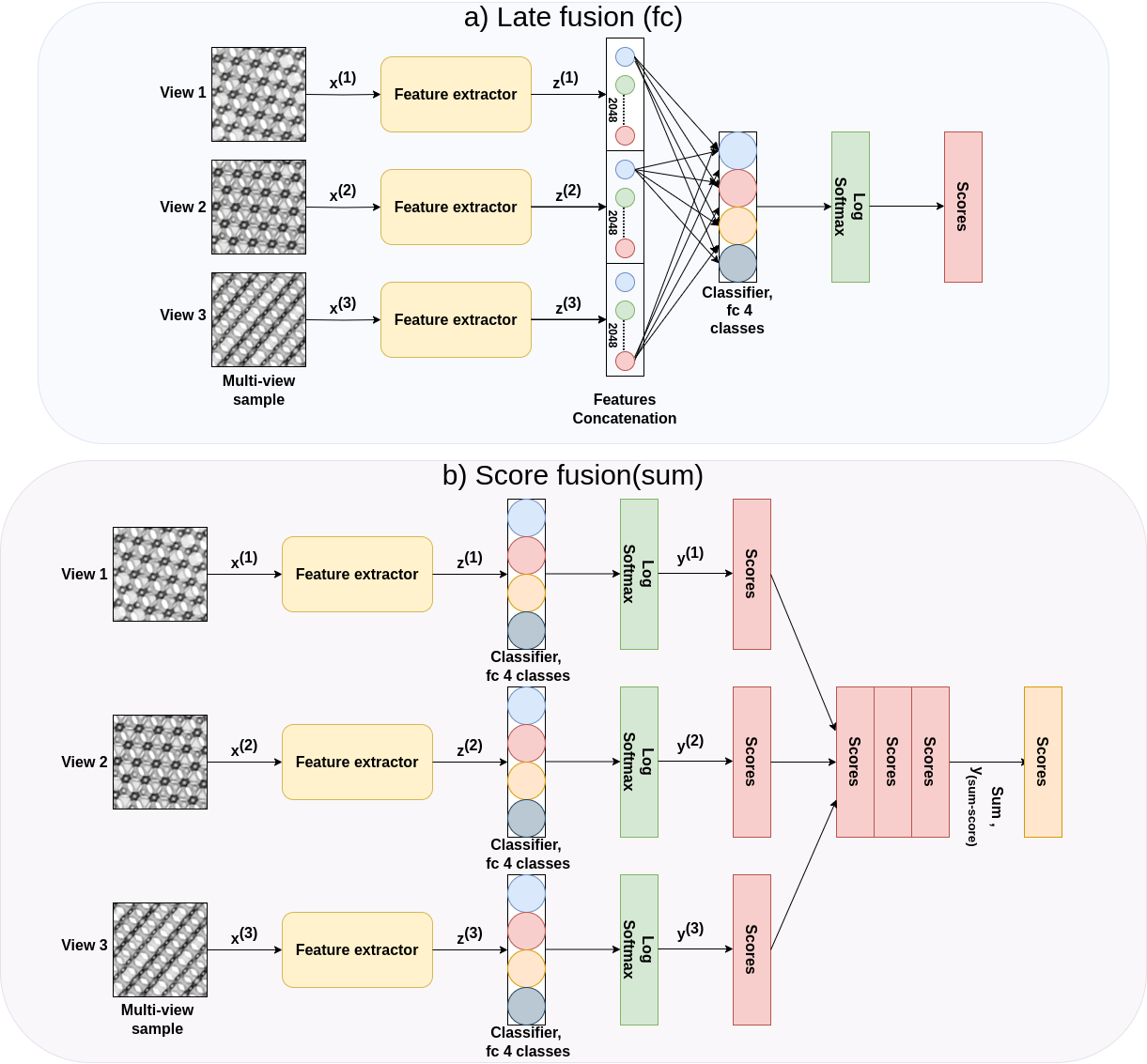
Biological membranes are vital for cellular functioning in both plants and animals. Understanding the 3D structures of these membranes is essential to comprehend their functionality. Due to their nanoscale size, these structures are studied using transmission electron microscopy, which provides 2D micrographs of 3D structures. These micrographs are challenging to analyse due to their similar appearance. The aim of this study was to classify 2D micrograph projections using convolutional neural networks (CNNs) and compare two classification strategies: one using a single image of the micrograph and the other employing images from different sections of the same micrograph. The CNNs were trained on projections generated through SPIRE software. The results demonstrated that CNNs utilising a multi-view approach, considering multiple views of the micrograph, outperformed those using a single-view strategy. However, more rigorous testing on a wide variety of real TEM specimens is necessary to determine the efficacy of models trained on software-generated projections to real TEM micrographs.
Code and documentation: GitHub repo.